Review Article
Volume 3, Issue 5
Insect Chemosensory Proteins against COVID-19
Jean-François Picimbon1,2,3*
1Taishan Scholar, Biotechnology Research Center, Shandong Academy of Agricultural Sciences, 202 Gongye North Road, Jinan, Shandong, 250100, China.
2Distinguished Professor, School of Bioengineering, Qilu University of Technology, 3501 University Road, Jinan, Shandong, 250353, China.
3National Expert of China, Science and Technology Service Platform of Shandong Academy of Sciences, Shandong Academy of Sciences Foreign Students Pioneer Park, QLUT-SDAS, Jinan, 250103, Shandong, China.
Corresponding Author :
Jean-François Picimbon
Tel: 0086-531-83175350;
Email: jfpicimbon@gmail.com & jfpicimbon@sdas.org
Received : Mar 15, 2024 Accepted : May 06, 2024 Published : May 13, 2024 Archived : www.meddiscoveries.org
Citation: Picimbon JF. Insect Chemosensory Proteins against COVID-19. Med Discoveries. 2024; 3(5): 1152.
Copyright: © 2024 Picimbon JF. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
This article describes a novel strategy to fight SARS-CoV-2 infection by employing tiny soluble binding proteins called “Odorant Binding Proteins” (OBPs), “Niemann-Pick Type C2 (NPC2s)”, and “Chemosensory Proteins” (CSPs). The possible interactions between these proteins and sensory receptors are not relevant to COVID-19. We concentrate on the interaction between the protein-binding site and C18 lipids in COVID. Extended lipid chains, like palmitic acid (C18), oleic acid (C18:1), and linoleic acid (C18:2) resemble the primary components of membrane viral particles. The virus’s surface is covered in C18 lipids. In light of this, OBPs’, NPC2s’ and CSPs’ capacity to interact with C18-lipids and transfer them to Degradative Enzymes (ODEs) may provide a novel molecular strategy for combating coronavirus infection, regardless of the course of viral mutation. In order to confine COVID virus variations in a hydrophobic pocket and/or produce viral particles devoid of an outer protective envelope, proteins with the ability to bind long lipid chains on viral surfaces or peplos may show great promise.
Introduction
Covid-19 (Coronavirus Disease 2019) is an infectious respiratory disease evoking a health crisis and a global pandemic such as we rarely see in human history linked to a new type of virus, along the lines of SARS (Severe Acute Respiratory Syndrome, 2002-2004) and MRES (Middle East Respiratory Syndrome: first identified in 2012 and still prevalent in 2024) [1-3]. The first severe acute respiratory syndrome coronavirus, also known as SARS or SARS-CoV-1, was the cause of the 2002-2004 SARS epidemic outbreak that sickened over 8,000 individuals from 30 different countries and territories and claimed at least 774 lives globally. This pales in comparison to the second wave, which, as of May 26, 2020, had 5,603,558 confirmed cases and 348,194 deaths worldwide. In Nov. 2002, the SARS-1 outbreak was initially detected in Foshan, Guangdong, China. The second contagion, known as SARS-2, first surfaced on December 8, 2019, when several patients in Wuhan, Hubei Province, China, started exhibiting symptoms resembling pneumonia (cough, fever, and dyspnea). Similar to SARS-1, SARS-2 is caused by a virus particle from the genus Coronaviridae, which is family to the Coronaviruses (CoVs) and comprises pleomorphic RNA viruses with peplomers resembling crowns [4]. SARS-CoV-2 can cause and spread an acute respiratory illness that can be extremely serious, or it can cause a respiratory illness of mild to moderate intensity that can heal on its own without the need for special treatment. The coronavirus family is diverse [5]. The majority of them make animals sick. Nonetheless, human illness is known to be caused by seven different types of coronavirus. This list of human coronavirus infections includes four that cause a mild upper respiratory illness similar to a cold, but MRES, SARS-CoV-1, and SARS-CoV-2 have the potential to be far more dangerous, and they have recently been linked to significant outbreaks of fatal pneumonia [6]. Numerous questions regarding the birth, origin and evolution of the different coronavirus strains are raised by this diversity in pathogenicity and transmissibility. The question of how SARS-CoV-1 could have disappeared so quickly while SARS-CoV-2 is still around five years after spreading is one that requires special attention because it still generates a staggering amount of mutant strains and variations that differ from the original virus in many ways (Table 1) [6-10].
Aiming for the “Spike”
Since all of the viral mutations that define novel strains of the virus occur at the level of surface protein S, or “Spike”, this protein is the primary focus of research against Covid-19. Spike protein is one of the essential components of the peplos, or viral envelope or shield. The virus has a thorny shape covered with spikes that makes it resemble a crowned king [11,12]. This spike-shaped glycoprotein on the viral membrane surface is how SARS-CoV-2 enters human cells. By fusing the SARS-CoV-2 envelope with the host cell membrane, the S protein binds to ACE2 receptors on human cell membranes to transfer infectious RNA there [12-15]. As a result, research on SARS-CoV-2 is accelerating in several areas, such as the investigation and identification of the molecular underpinnings of the ACE2-S interaction and membrane fusion [15-20].
In response to numerous mutations on the spike protein of the SARS-CoV-2 [21-23], the major biotechnology companies developed approximately forty vaccines for the purpose of developing viral products and conducting clinical trials, logistics, and manufacturing across multiple countries and country associations (Table 2). The ten Covid vaccines listed in Table 2 are those that have been given the go-ahead for emergency or widespread use by at least one strict regulatory body accredited by the World Health Organization (WHO): Convidecia, Covaxin, CoronaVac, Janssen (Johnsson & Johnsson), Moderna (USA), Novavax, Oxford-AstraZeneca (UK), Pfizer-BioNTech, SanofiGSK, and Sinopharm (BIBP, Beijing, China). The WHO is currently evaluating Abdala (CIGB-66, Cuba), Corbevax (Texas Children’s Hospital, USA), COVIran Barekat (Shifa Pharmed Industrial Group, Iran), SCB-2019 (Clover Biopharmaceuticals, USA Inc., Ireland Ltd), Sinopharm WIBP (WIBP-CorV, Sinopharm, China), Sputnik V (Gam-COVID-Vac, Gamaleya Research Institute of Epidemiology and Microbiology, Russia), and Zifivax (ZF2001 or ZFUZ-VAC-2001, Anhui Zhifei Longcom, China), among the other anti-Covid vaccines. The fact that there are so many vaccines and technologies available only suggests that there are as many potential vaccinations as there are variants on a spike, but the best course of action is still up for debate.
If vaccination is the only treatment for Covid, then given the number of patients who are against vaccinations, we must also take into account the possibility that Covid and Spike will continue to evolve and mutate (as evidenced by the number of variants discovered after five years), eventually eluding our supply of vaccines and/or depleting our ability to continuously try and modify the vaccine to account for the new mutation. The history of Covid virus is another thing to think about [24]. SARS-CoV-2’s exact chain of animal-to-human transmission is still unknown, but a zoonotic origin, in which the virus started in bats before moving on to an intermediary host (probably a pangolin) where it underwent a mutation and eventually spread to humans, is one of the most likely scenarios for the virus’s evolutionary origin. Whatever the history, one crucial aspect of the current scenario is the efficient infection and replication ability of SARS-CoV-2, which has led to the emergence of viral variants that are more contagious and have varying degrees of pathogenicity than the original virus, raising concerns about their containment. The severity of illness and mortality caused by SARSCoV-2 infection is decreasing due to the availability of vaccines, one after the other over multiple adaptations, repetitions, and injections; however, the virus’s extinction is not expected, predictable, and/or imminent. Most of the current SARS-CoV-2 neutralizing antibodies were unable to neutralize Omicron [25]. Regarding this, humoral immune escape was a defining feature of Covid re-emergence, which has emphasized the importance of tracking SARS-CoV-2 evolution globally.
Furthermore, since bats are flying mammals rather than birds and are thought to be the original host of all β- and α-coronaviruses, as well as human coronaviruses, and many more, this is also required. Most of all bat chiropteran families have tested positive for many various coronaviruses. Ninety percent of the 100 distinct viruses that cause human diseases worldwide on hotspots have been identified in bats. It is anticipated that bats harbor over >3200 coronaviruses, the vast majority of which are currently unidentified. These include pathogens that cause high-consequence infectious diseases like the filoviruses that cause Ebola and Marburg, paramyxoviruses that cause Nipah and Hendra, and all of these human-emerging MERS-CoV, SARS-CoV, and SARS-CoV-2 variants. These variants share the same human cellular receptor molecule as SARSCoV-2, and as such, they may present a greater risk of emerging from bats into humans in the future [26,27]. Do we have the resources and the technology to keep up the fight and consistently adjust to a new bat scenario by producing a large quantity of anti-spike vaccines for every population?
Numerous other animals, in addition to bats, are also potential carriers of coronaviruses, even though bats harbor a higher number of zoonotic illnesses and newly emerging coronaviruses than any other mammalian taxa. The coronavirus threat may arise from coronaviruses that have been found to be harmful to companion animals, livestock, and lab animals. Human coronavirus spillover is a concern for a wide range of domestic and wildlife species, including Canis lupus, Felis catus, felines, wild-apes, rhesus macaques, chimpanzees, African green monkeys, gorillas, civets, ferrets, chickens, ducks, minks, rodents, murids, pigs, ungulates, camels, deers, horses, sheeps, bovines, and raccoons. The majority of these animals coexist with humans in increasingly close quarters as a result of deforestation and urbanization throughout the world. Since the majority of these animals are living in increasingly close proximity to human habitats, this poses a widespread issue for human society. SARS-CoV-2, the “human” version of the virus, has infected a wide range of animal species. The possibility of the pathogen to spread back to humans through arises from the possibility of interspecies transmission producing new animal reservoirs where the virus can proliferate or endure for extended periods of time [28,29]. Since most of these animals share the same ecosystem with humans, we come into constant contact with them—all the more so in this era of climate change and unpredictable seasonal growth. This will increase the likelihood of viruses spreading from humans to animals and vice-versa [29,30]. Mutations on the spike protein can multiply in all forms of zoonosis and/or reverse zoonosis, which can be especially harmful to humans and great apes, for example. As our closest living relatives and/or coexisting with humans in zoos or reserves, great apes are a taxonomic group that is vulnerable to human spillover. Every great ape species faces extinction. With our arsenal of vaccines (see Table 2), will we be able to save them? If the bat spike develops into an ape, a pig, a chicken, or a cat spike that is especially virulent for humans, are we prepared to modify our vaccinations? Are our tools worthless in the long run if spike mutations are the result of perpetual environmental fluctuations and climate change? In order to combat COVID-19 and its future offspring, we probably need to continue looking for a more universal solution than treating spike, given the large number of potential hosts and coronaviruses that can cause a novel human disease.
Aiming for the “Crown”
The lipid crown should be the target. The main components of the SARS-CoV-2 particle are proteins and fatty acid lipids. The lipid bilayer of the virus is made up of phospholipids (phosphatidylcholine, phosphatidylethanolamine, and sphingomyelin), glycolipids, cholesterol, and generally glycerol, as well as fatty acid lipids such as palmitic acid (C18), oleic acid (C18:1), and linoleic acid (C18:2). When it comes to lipids, there is threefold interest: 1) the SARS-CoV-2 envelope is distinct from the host cells; 2) the virus’s exposed procoagulant lipids are accessed because oral rinses disrupted them in vivo; 3) the virus picks its lipid envelope from the host’s cell’s membrane [31-33]. The chemical makeup of the virus membrane plays a crucial role in the virus’s ability to fuse and enter target cells, indicating that effective SARS-CoV-2 infection may depend on the maintenance of a functional lipid bilayer structure [32,33]. These fats, which are recognized as crucial plasma lipoprotein surface components, have a major impact on the development of SARS, viral replication, the viral infectious cycle, and COVID-19 [34-43]. Thus, targeted lipid-based treatments for SARS-CoV-2 infection ought to be very effective. One of the virus’s true chimal characteristics is its lipidic composition [44,45], and further research could be done on the supersingular COVID-19 regulation, which is based on lipid transport proteins like “Pheromone-Binding Proteins” (PBPs), “ChemoSensory Proteins” (CSPs), and Niemann-Pick Type C2 proteins (NPC2s) from insects.
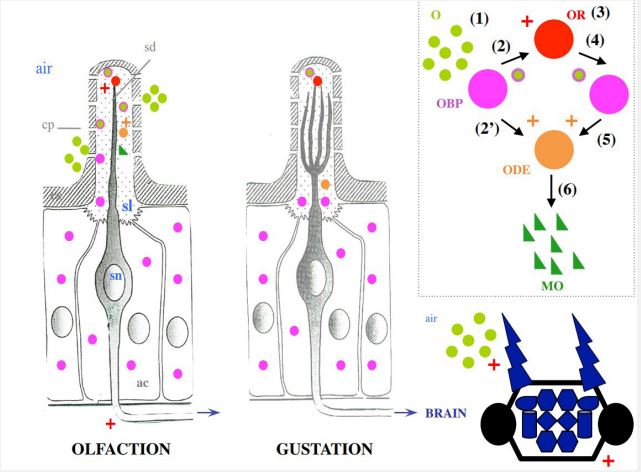
As extensively described in the study of moths, the olfactory cells observed in electronic microscopy exhibit a clear functional structure. This is a system of tubular pores (where odorous molecules enter) that reflects the surrounding air to the Olfactory Receptors (ORs) [46]. The chemical and physical structures of odorous molecules, such as the sexual pheromones of moths, are typically very hydrophobic, with long carbon chains leading to aldehyde, alcohol, or acetic acid, serving as the only hydrophilic polar unit of the molecule. Similar to fatty acid lipids, odorous molecules close to the cells inside the olfactory cilia (sensilla) must pass through an aqueous environment, a naturally occurring hydrophilic barrier, the sensory lymph node of insects, in order to reach ORs and the dendrites of olfactory receptor neurons (Figure 1).
The various senses of taste and smell preserve features such as tubular pores, sensory lymph surrounding dendrites, cilia structure, and receptors isolated from airborne volatiles by an aqueous medium. This typical structure is also present in contact chemosensory sensilla of insects (Figure 1) [47]. A typical model of the molecular basis of smell and taste reception in insects looks like this: lipid chains and odorous molecules were able to become soluble due to notably high concentrations of small soluble proteins in the lymph, known as Odorant-Binding Proteins (OBPs), that enhanced and facilitated their transport to the ORs and/or different metabolic enzymes like Cytochrome oxidases P450 (CYPs) and Odor-Degrading Enzymes (ODEs) [48- 49]. “OR, ODE, OBP” is widely accepted to be the cornerstone of the biochemical basis for the detection of olfactory cues at the periphery of the Central Nervous System (CNS) (see Figure 1), though this is still up to debate [50].
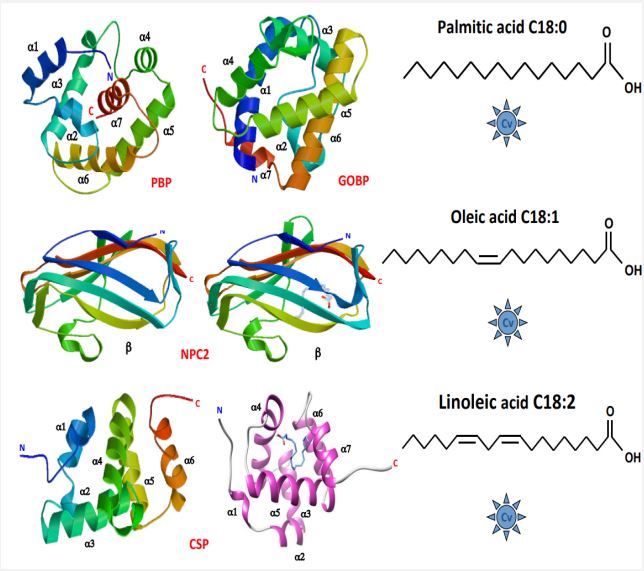
Three protein types—OBP, CSP, and NPC2—have been linked to a possible role in olfaction and/or taste in insects [51]. In the moth system, OBP and CSP have been well documented [52- 55], whereas NPC2 is thought to mediate chemical communication in ants exclusively [56]. However, the biochemical and binding characteristics of these protein families are of greater interest to COVID-19 than any potential roles these proteins may play in odor signal transduction (Figure 2). For PBP identified in moths, a hydrophobic pocket is formed from four central α-helices, leaving the N- and C-termini free and exposed to the entry and/or release of regulatory ligands [57]. The PBP links not only to pheromone products but also to palmitic acid (C18) [58], which is particularly interesting for the fight against COVID-19 since C18 is a major component of the lipid layer of SARS-CoV-2 (Figure 2). Comprising six α-helices, the CSPs have a similar function. With an extremely hydrophobic internal binding pocket, the CSP structure is suitable for extraction and transportation of lipid chains such as linoleic acid (C18:2) [59]. This transfer of C18:2 also provides an intriguing connection to the Covid peplos structure (Figure 2). Lastly, NPC2 is known to interact in worker ants with oleic acid (C18:1) [60], which may have a direct bearing on controlling the Covid virus. These interactions (PBP-C18, NPC2-C18:1, and CSP-C18:2) are a critical link between insect binding protein families and SARS-CoV-2 surface lipids (Figure 2).
Stricking SARS-CoV-2’s lipid degradation
The hypothesis regarding the various mechanisms of action of OBP, CSP, or another NPC2 in the therapeutic treatment of COVID-19 has been described by Picimbon (2023) [61]. The injection of OBP may form a hydrophobic stock around host cells, which could have six significant effects on the infection mechanism: 1) inhibition of transmembrane serine protease 2 (TMPRSS2) binding site, 2) inhibition of the Angiotensin-Converting Enzyme 2 (ACE2) binding site, 3) destabilization of the SARSCoV-2 envelope due to the attachment of the OBP to lipids on the surface of viral particles, 4) repair of host cell membranes by afflux of long lipid chains of the fatty acid type C18, 5) interaction with ODE and CYP metabolic enzymes, which break down the lipids that make up the SARS-CoV-2 envelope, and 6) destruction of the viral ARN (by therapeutic molecules like interferon INFα, which targets the virus without peplos) following the breakdown of the lipid layer of the viral capsid [61].
At this time, there is no proof that the COVID-19, or more precisely the virus SARS-CoV-2, is related to PBP, CSP, or NPC2 complexed with ODE, although there are a few potential signs of interactions:
• The synthesis of specific OBP in the saliva and peripheral nervous system of Aedes aegypti (yellow fever mosquito) is caused by the dengue virus
• Certain coronavirus variants stimulate the production of Mouse Urinary Proteins (MUPs)
• The SARS-CoV-2 significantly alters the activity of CYP involved in the metabolism of fats
• The C18 lipid chains that PBP, CSP, and NPC2 interact with have been shown to be essential for viral replication and fusion with the host cell membrane ([61]).
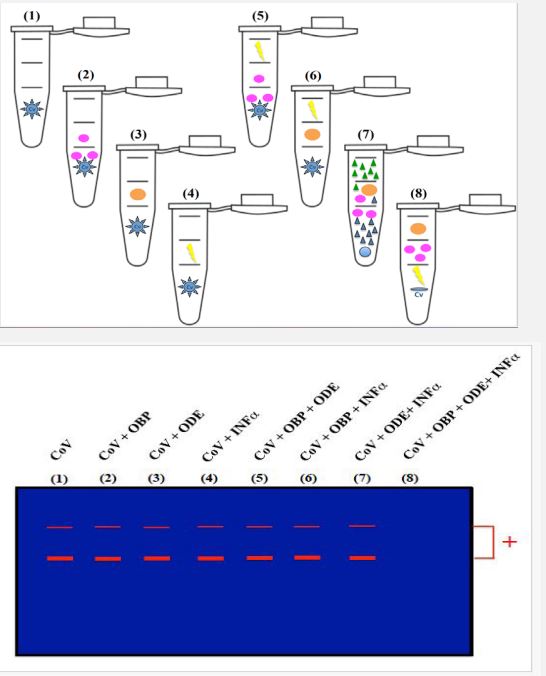
Here, we may concentrate on what is known and practical for SARS-CoV-2 using PBP, CSP, and/or NPC2 (see Figure 3). There is no evidence of a direct interaction between PBP, CSP, and NPC2 and SARS-CoV-2, but there is strong evidence of a direct interaction between PBP, CSP, and NPC2 and the lipid constituents of the virus peplos [see 58-61]. It’s a new avenue for human research and clinical medicine without implying that this approach is directly applicable at this time and/or place. However, it is possible to test the insect binding proteins on the virus at this point (Figure 3). In bacterial systems, OBP, CSP, and NCP2 are easily expressed in large quantities for the analysis of functional properties and the resolution of three-dimensional structures. The initial crucial and non-neglectable step in the process is sub-cloning in an Escherichia coli protein expression system, which yields soluble recombinant proteins. Then, biochemical methods (selective affinity chromatography, fusion His-Tag protein, fast protein liquid chromatography, and/or gel filtration) are to be used to extract and purify these small soluble proteins, ranging in size from 10 to 19 kDa, from insect tissues bruised [see 51-61]. Following the production of the protein and the addition of CYP and/or ODE as well as interferon, further techniques include a simple PCR and/or a COVIDtest for the detection of SARS-CoV-2. There shouldn’t be any signals on gel electrophoresis and/or conventional Polymerase Chain Reaction (PCR) in real-time PCR assay for the detection of SARS-CoV-2 if PBP, CSP, NPC2, or the three proteins together, have been able to effectively trap the viral lipids and deliver them to metabolic enzymes [62,63] (Figure 3). Better than PCR alone, Whole Genome Sequencing (WGS) can be necessary to identify novel variants in the populations. RT-PCR genotyping assays, which can be used as a substitute for WGS, can identify the distinctive set of mutations present in the genomes of SARS-CoV-2 variants. These genotyping tests are usually made to distinguish between original strain and mutant sequence at a hot spot [64]. A specific variant may be present in the sample if a variant mutation causes the target amplification in some diagnostic COVID-19 detection assays to fail. Consequently, after being exposed to OBP, ODE, and INFα, we could use PCR, realtime PCR, WGS, and/or genotyping experiments to determine whether the virus is still present. Regardless of the variant, this combination OBP+ODE+INFα could make the target amplification fail (see Figure 3).
C18:2-CSP and COVID-19
As an alternative, we could access the virus’s integrity using protein separation and Western blot. It is most likely crucial to choose a specific antigen as the target for SARS-CoV-2 immunodetection when using the protein detection method [65]. Both Spike and N proteins have been demonstrated to be essential structural proteins with high immunogenicity in this context, and they both have have the potential to be the main targets of anti-Covid immunoassay. After the lipid capsule is broken down by OBP+ODE treatment, we should be able to separate protein S from the rest of the virus (Figure 3). This could be useful for carrying out additional biochemical research on the configuration of isolated Spikes. When attemting to control Covid via a control on Spike, it is important to take into account the distinct conformational states of the SARS-CoV-2 spike protein [66,67]. The conformational transitions of Spike are essential for the S protein structure to associate with ACE2, engage its receptor binding domain with ACE2, activate S through proteolysis, endocytosis, and membrane fusion; which is a crucial mechanism of SARS-CoV-2 entry into host-cells and infection [68]. Crucially, it has been discovered that fatty acid lipids—specifically, linoleic acid (C18:2)—control the conformational changes of Spike [69- 71]. The structure of SARS-CoV-2 spike S glycoprotein, determined by cryo-electron microscopy at 2.85 Å, shows that C18:2 is tightly bound by the receptor binding domains in three composite hydrophobic binding pockets [71]. This finding highlights the need to investigate the interactions between lipids and spike—more especially, C18:2 and S—that regulate the coronavirus, and consequently, the potential involvement of CSP in this mechanism. It would be especially effective to use CSP’s ability to transfer C18:2, regardless of the coronavirus species. MERS-CoV and SARS-CoV also seem to contain a very similar hydrophobic pocket [71]. A locked S conformation is stabilized by C18:2 binding, which reduces the ACE2 interaction in vitro. C18:2 supplementation works in concert with remdesivir (Veklury) in host cells to inhibit the replication of the virus [71,72]. The structure of the spike protein, in addition to the virus’s lipid surface, builds a direct link between C18:2, CSP, and Covid. This opens the door for intervention strategies that concentrate on lipid binding, transport, and/or transfer, particularly C18, C18:1, and C18:2, by SARS-CoV-2. It might be particularly interesting to look for differences in the conformational changes and different conformational intermediates of spikes after being exposed to OBP, CSP, and/or NPC2 in addition to membrane fusion. Through the use of Nuclear Magnetic Resonance (NMR), amide hydrogen/deuterium exchange mass spectrometry, and biochemical analysis of spike conformations, it may be possible to determine how CSP affects spike folding and, in turn, inhibits the virus’s ability to fuse with the host cell membrane [73-75].
Table 1: A chronology and diversity of SARS-CoV-2 variants and strains. The number in brackets is the number of spike mutations.
| Variant (n) | Subvariant | Emergence | Country | Contagiousness | Symptoms | Vaccines |
|---|---|---|---|---|---|---|
| Alpha (3) | B.1.1.7 | Nov-20 | Great Britain | VOC † | Fever, chills, cough, shortness of breath, difficulty breath- ing, fatigue, muscle ache, body ache, headache, loss of taste and/or smell, sore throat, runny nose, nausea, vomiting, diarrhea, severe pneumonia^ |
++ |
| Beta (5) | B.1.351 | Dec-20 | South Africa | ≥ 50%* | Lightweight√ | +/- |
| Delta (4) | B.1.617.2 | Nov-20 | India | Predominant ------------------ In the World |
Headache, sore throat, runny nose, fever | + |
| Epsilon (2) | B.1.427/B.1.429 -------------------- Pango |
Sep-20 | United States | Highly transmissible | Adjusted ------------- T-cell^^ |
+ |
| B.1.616 | Feb-21 | France | ||||
| Eta (3) | B.1.525 | Dec-20 | Nigeria | Fever, dry cough, tiredness | ||
| Gamma (5) | P.1 | Dec-20 | Brazil | Increased | Increased ------------- coryza------------ myalgia |
++ |
| (8) | B.1.640 | Sep-21 | Congo | |||
| XD | Jan-22 | France | Delta-like | Delta-like | ||
| XF | Jan-22 | Great Britain | Omicron-like | Omicron | ||
| Iota (3) | B.1.526 | Dec-20 | United States | ≥15-25%* | Mild | - |
| (2) | B.1.526.1 | Oct-20 | United States | |||
| (2) | B.1.526.2 | Dec-20 | United States | |||
| Kappa (4) | A.23.1 | Dec-20 | Great Britain | VOI | Infection rashes, high fever, cough, runny nose, red and watery eyes |
- |
| A.27 | Dec-20 | N/A | High | High | ||
| (3) | A.28 | Dec-20 | N/A | |||
| (4) | `B.1.1.7 | Jan-21 | Great Britain | Increased | Increased | |
| (4) | `B.1.1.7 | Jan-21 | Great Britain | Increased | Increased | |
| (6) | B.1.351 | Dec-20 | South Africa | Increased | Increased | |
| (6) | B.1.351 | Jan-21 | N/A | Increased | Increased | |
| B.1.617.1 | Dec-20 | India | Increased | Increased | ||
| B.1.617.3 | Feb-21 | India | ||||
| B.1.620 | Feb-21 | N/A | ||||
| B.214.2 | Dec-20 | N/A | ||||
| (2) | C.16 | Oct-20 | N/A | |||
| Lambda (3) | C.37 | Dec-20 | Peru | VOI/VOC | High fever, continuous cough, blocked nose, fatigue, muscle pain, ------------------------------------------------------------------------- body ache |
+/- |
| -6 | AY.4.2 | Jun-21 | Great Britain | Increased | Similar | |
| -3 | B.1.1.318 | Jan-21 | N/A | |||
| -5 | B.1.617.2# | Jun-21 | Great Britain | |||
| -5 | B.1.617.2# | Apr-21 | India | |||
| -5 | B.1.617.2# | Apr-21 | India | |||
| -5 | B.1.617.2# | Apr-21 | India | |||
| C.1.2 | Jun-21 | South Africa | Increased | Increased | ||
| Mu (5) | B.1.621 | Jan-21 | Colombia | Very high | Shortness of breath, sore throat, fever, shivering, fatigue, headache, runny nose, nausea, vomiting, diarrhea |
+ |
| Omicron | BA.1 | Nov-21 | Bostwana | ≥1 M cases | Runny nose, cough, fever, headache, muscle ache, mild/ severe fatigue, sore throat, pain, sneezing, hoarse voice |
++ |
| (≥ 30) | South Africa | |||||
| BA.2 | Nov-21 | South Africa | Increased | Increased | ||
| -1 | BA.2+ | N/A | N/A | |||
| BA.2.75 | May-22 | India | VOI (unclear) | Baseline | ||
| -6 | BA.2.86 Pirola | N/A | N/A | VOI (unclear) | Und | ++ |
| -3 | BA.2.3.20 | N/A | N/A | |||
| BA.3 | Nov-23 | South Africa | ||||
| -3 | BA.4 | Jan-22 | South Africa | |||
| -3 | BA.5 | Feb-22 | South Africa | |||
| -2 | BF.7 | N/A | N/A | |||
| -3 | BN.1 | N/A | N/A | |||
| -2 | BQ.1 | N/A | N/A | |||
| -1 | B.1.1.529 | N/A | N/A | |||
| -2 | B.1.1.529 | N/A | N/A | Increased | ||
| -2 | CH.1.1 | N/A | N/A | |||
| -3 | DV.7.1 | VUM | ||||
| EG.5 (Eris) | ++ | |||||
| XAK | Jun-22 | Germany | ||||
| -1 | XAY | N/A | N/A | |||
| -2 | XBB | N/A | N/A | Increased | Increased | |
| -3 | XBB.1.5-like | N/A | United States | VOI (Baseline) | Baseline | |
| -3 | XBB.1.16 | N/A | N/A | |||
| -4 | XBB.1.16.6 | N/A | N/A | VOI (dominant) | Baseline | |
| -2 | XBC | N/A | N/A | |||
| Theta (4) | P.3 | Jan-21 | Philippines | VOI | % Hosp ^^^ | - |
| Zeta (3) | P.2 | Jan-21 | Brazil | VOI | Baseline | ++ |
| -4 | AV.1 | Mar-21 | Great Britain | |||
| -4 | AT.1 | Jan-21 | Russia | |||
| -2 | B.1.1.519 | Nov-20 | Mexico | |||
| -3 | C.36 | Dec-20 | Egypt | |||
| -6 | P.1 | Feb-21 | Italy |
VOC: Variant of Concern; VOI: Variant of Interest; VUM: Variant under Monitoring; Hosp: Hospitalization for severe acute pneumonia syn-
drome. *: means in % more contagious than the initial strain of the virus (SARS-CoV-2, Oct-Dec 2019, Wuhan, Hubei Province, China); #: is distinct
due to the spike mutation (+K417N, +E484X, +Q613H, and +Q677H). `: +L452R or +S494P mutation. N/A stands for unclear origin with respect to
time and place. Baseline means the impact on transmissibility or immunity is similar to baseline. Compared to the new original variant, increased
indicates a greater impact on transmissibility or immunity; †: De-escalated variants (it is no longer in circulation, it has been in circulation for a
long time without having an effect on the overall epidemiological situation, or scientific evidence shows that it is not linked to any properties that
should be of concern); Vaccines (Pfizer, Moderna, Johnson & Johnson): - not efficient (more resistant to neutralizing antibodies); + efficient; ++
very efficient. The subvariant or lineage gaining a higher and more dangerous profile is indicated by the name in red.
Data are from Yale Medicine (Sept. 1st 2023; yalemedicine.org) and European Centre for Disease Prevention and Control (ECDC, Agency from EU,
Dec. 1st 2023; ecdc.europa.eu).
√Citing Jonathan Ball, Professor of Molecular Virology at Nottingham University (2021)
^Niv et al. (2023) [7]
^^Plummer et al. (2022) [8]
^^^ Haw et al. (2022) [9
Table 2: List of vaccines against Covid-19 infection approved for emergency or full use (WHO, World Health Organization; en.wikipedia. org).
| Vaccine | Brand | Vaccine | Country | Efficiency | Secondary effects |
|---|---|---|---|---|---|
| name | type | ||||
| Convidecia | AD5-nCOV | Viral vector |
China & Malaysia,
Mexico, Pakistan |
(1) 66% | Pain, redness, induration, swelling, itchy (injection site) |
| CoronaVac | Sinovac |
Inactivated Covid virus |
China (3) | 98% |
Injection site redness,
pain, headache, nausea,
flu-like condition, lymphadenopathy, diarrhea, weakness |
| Covaxin | BBV152 |
Inactivated Covid virus |
India (1) | 64% |
Mild fever, pain, injection
site tenderness, fatigue,
headache, myalgia, *malaise, *pyrexia |
| Janssen | Jcovden | Viral vector | Netherlands | (1) 66% |
Flu-like, fatigue, fever,
headache, muscle aches,
thrombosis*, thrombocytopenia* |
| J & J | & Belgium | ||||
| Moderna | Spikevax | mRNA-based | United States | (2) 94% | Injection site pain, fever, headache, strong fatigue∫ |
| Novavax | Nuvaxovid | Subunit Covid | United States & CEPI | (2) 90% |
Fever, headache, nausea,
muscle pain, joint pain,
tiredness, *ana- phylaxis, *paresthesia, *hypoesthesia, *pericarditis |
| Covovax | |||||
|
Oxford-As- traZeneca |
Covishield | Viral vector | Great Britain & Sweden | (2) 81% | Injection site pain, nausea, headache, |
| Vaxzevria | anaphylaxis* | ||||
|
Pfizer-BioN- Tech |
Comirnaty | mRNA-based | United States & Germany | (2) 97% | Injection site pain, fatigue, |
| headache, | |||||
| allergy* | |||||
| Sanofi-GSK | VidPrevtyn Beta |
Protein subunit | France & Great Britain | (2) 58% |
Tiredness, nausea, diarrhea,
swelling, pain, redness
(injection site), sore arm, light flu |
|
Sinopharm BIBP |
BBIBP-CorV |
Inactivated Covid virus |
China | (2) 90% |
Tiredness, chills, fever,
dizziness, headache,
swelling, pain, redness (injection site), |
| & UAE | sore arm, myalgia, general lethargy |
*Rare cases. ∫: The signs and symptoms I personally encountered 24 hours following the arm deltoid muscle injection. The number of doses
required to increase the level of protection against SARS-CoV-2 to the specific percentage is shown in brackets. CEPI: Coalition for Epidemic
Preparedness Innovations (Wellcome Trust, Bill & Melinda Gates Foundation, India, Norway, EU, UK, and World Economic Forum; Headquarters,
Oslo, Norway).
J & J: Johnsson & Johnsson (one dose, rare adverse event: blood clotting)
CDC-Centers for Disease Control and Prevention (cdc.gov)
Conclusion
MERS-CoV, SARS-CoV, the survival of SARS-CoV-2 approximately five years after it first appeared globally, the threat posed by numerous variants (variation, version, descent, “nationality”, Greek name, number, origin, line, lineage, branch, split, strain, derivation, mutation, substitution, variance, or sub-viral peplos variance), the likelihood that the coronavirus will continue to mutate and that a new one will emerge, our increasing proximity to animals, the potential for reverse-zoonosis, and the side-effects of immunizations force us to think of novel approaches to coronavirus infection management. It turns out that, in the case of SARS-CoV-2 the lipid composition of the peplos is not dependent on the strain or variation of the virus. Fatty acid lipids, like linoleic acid, are especially significant for the infection and the replication of the virus as well as for the conformational change of the spike protein, which is one of the main structural proteins of the virus and its fusion with membrane cells. This makes it possible for us to propose using insect binding proteins—specifically PBPs, CSPs, and NPC2s— to combat COVID-19 illness. Due to their relationships to C18 lipids and the breakdown of these lipids by metabolic enzymes, PBPs, CSPs, and NPC2s represent an important class of proteins for future studies on COVID-19 and novel therapeutic challenges. These treatments, which use proteins derived from insects, have a great deal of potential to replace vaccinations and/or provide comprehensive defense against all coronavirus kinds.
Declarations
Funding: Overseas high-level talent & Taishan scholar title (#NO.tshw20091015) - National Expert title (China; #G2022023033L).
Conflict of interest: N/A
Ethics committee: N/A
Consent: N/A
Author’s contribution: The named author (JFP) has granted approval for the article’s final publication and satisfies the requirements set forth by the International Committee of Medical Journal Editors. JFP also assumes full responsibility for the working concept developed here.
References
- COVID-19 Resource Center. COVID-19 content collection. THE LANCET. https://www. who. int/emergencies/diseases/novelcoronavirus-2019 . WHO: Coronavirus disease (COVID-19) outbreak; Cell Press Coronavirus Resource Hub. Elsevier’s Novel Coronavirus Information Center, Elsevier Coronavirus Research Hub, 2019; Harvard Health; https://www. health. harvard. edu/diseases-and-conditions/coronavirus-resource-center, December 4th 2023.
- Chery JD, Krogstad P. SARS: the first pandemic of the 21st century. Pediatr Res. 2004; 56: 1-5.
- World Health Organization (29 August 2023). Disease Outbreak News; Middle East respiratory syndrome coronavirus (MERSCoV) - Saudi Arabia; https://www.who.int/emergencies/disease-outbreak-news/item/2023-DON484.
- Narayanan SA, Jamison DA, Guarnieri JW, Zaksas V, Topper M, et al. A comprehensive SARS-CoV-2 and COVID-19 review, Part 2: host extracellular to systemic effects of SARS-CoV-2 infection. Eur J Hum Genet. 2024; 32: 10-20.
- Víkovski P, Kratzel A, Steiner S, Stalder H, Thiel V, et al. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021; 19: 155-170.
- Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019; 17: 181-192.
- Niv Y, Kuniavsky M, Bronshtein O, Goldschmidt N, Hanhart S, et al. Difference between COVID-19 Alpha Variant B. 1. 1. 7 and the original virus in gastrointestinal symptoms and mortality: does a negative correlation exist? Isr Med Assoc J. 2023; 25:453-455.
- Plummer JT, Contreras D, Zhang W, Binek A, Zhang R, et al. US Severe Acute Respiratory Syndrome Coronavirus 2 Epsilon Variant: highly transmissible but with an adjusted muted host T-Cell response. Clin Infect Dis. 2022; 75: 1940-1949.
- Haw NJL, CaÒal EMR, Zuasula J Jr, Loreche MJ, Bernadas J. Epidemiological characteristics of the SARS-CoV-2 Theta variant (P. 3) in the Central Visayas region, Philippines, 30 October 2020-16 February 2021. Western Pac Surveill Response J. 2022; 13: 1-3.
- Carabelli AM, Peacock TP, Thorne LG, Harvey WT, Hugues J, et al. SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat Rev Microbiol. 2023; 21: 162-177.
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020; 367: 1260-1263.
- Mittal A, Manjunath K, Ranjan RK, Kaushik S, Kumar S, et al. COVID-19 pandemic: Insights into structure, function, and hACE2 receptor recognition by SARS-CoV-2. PLoS Pathog. 2020; 16: e1008762.
- Wang M, Zhao R, Gao L, Gao X, Wang D, et al. SARS-CoV-2: Structure, biology, and structure-based therapeutics development. Front Cell Infect Microbiol. 2020; 10: 587269.
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016; 3: 237-261.
- Ke Z, Oton J, Qu K, Cortese M, Zila V, et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 2020; 588: 498-502.
- Ou X, Liu Y, Lei X, Li P, Mi D, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune crossreactivity with SARS-CoV. Nat Commun. 2020; 11: 1620.
- Shang S, Ye G, Shi K, Wan Y, Luo C, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020; 581: 221-224.
- Plante JA, Liu Y, Liu J, Xia H, Johnson BA, et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2020; 592: 116-121.
- Tang T, Bidon M, Jaimes JA, Whittaker GR, Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020; 178: 104792.
- Beyerstedt S, Casaro EB, Rangel B. COVID-19: angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. Eur J Clin Microbiol Infect Dis. 2021; 40: 905-919.
- Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021; 19: 409-424.
- Chen KK, Huang DT, Huang LM. SARS-CoV-2 variants – Evolution, spike protein, and vaccines. Biomed J. 2022; 45: 573-579.
- Markov PV, Ghafari M, Beer M, Lythgoe K, Simmonds P, et al. The evolution of SARS-CoV-2. Nat Rev Microbiol. 2023; 21: 361-379.
- Pagani I, Ghezzi S, Alberti S, Poli G, Vicenzi E. Origin and evolution of SARS-CoV-2. Eur Phys J Plus. 2023; 138: 157.
- Cao Y, Wang J, Jian F, Xiao T, Song W, et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 2022; 602: 657-663.
- Irving AT, Ahn M, Goh G, Anderson DE, Wang FA. Lessons from the host defences of bats, a unique viral reservoir. Nature 2021; 589: 363-370.
- Anthony SJ, Johnson CK, Greig DJ, Kramer S, Che X et al. Global patterns in coronavirus diversity. Virus Evol. 2017; 3: vex012.
- Ghai RR, Carpenter A, Liew AY, Martin KB, Herring MK, et al. Animal reservoirs and hosts for emerging alphacoronaviruses and betacoronaviruses. Emerg Infect Dis. 2021; 27, 1015-1022.
- Madhusoodanan J. Animal reservoirs—Where the next SARSCoV-2 variant could arise. JAMA 2022; 328: 696-698.
- Ford JD, Zavaleta-Cortijo C, Ainembabazi T, Anza-Ramirez C, Arotoma-Rojas I et al. Interactions between climate and COVID-19. Lancet Planet. Health 2022; 6: e825-e833.
- Saud Z, Tyrrell VJ, Zaragkoulias A, Protty MB, Statkute E et al. The SARS-CoV2 envelope differs from host cells, exposes procoagulant lipids, and is disrupted in vivo by oral rinses. J Lipid Res. 2022; 63: 100208.
- Huarte N, Carravilla P, Cruz A, Lorizate M, Nieto-Garai JA et al. Functional organization of the HIV lipid envelope. Sci Rep. 2016; 6: 34190.
- Alketbi EH, Harndy R, El-Kabalaway A, Juric V, Pignitter M et al. Lipid-based therapies against SARS-CoV-2 infection. Rev Med Virol. 2021; 31: e2214.
- Lorizate M, Kräusslich HG. Role of lipids in virus replication. Cold Spring Harb Perspect Biol. 2011; 1: a004820.
- Heaton NS, Randall GR. Multifaceted roles for lipids in viral infection. Trends Microbiol. 2011; 19: P368-375.
- Ketter E, Randall G. Virus impact on lipids and membranes. Annu Rev Virol. 2019; 6: 3149-340.
- Dias SSG, Soares VC, Ferreira AC, Sacramento CQ, FintelmanRodrigues N et al. Lipid droplets fuel SARS-CoV-2 replication and production of inflammatory mediators. PLoS Pathog. 2020; 16: e1009127.
- Mandala VS, McKay MJ, Shcherbakov AA, Dregni AJ, Kolocouris A et al. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat Struct Mol Biol. 2020; 27: 1202-1208.
- Theken KN, Tang SY, Sengupta S, FitzGerald GA. The roles of lipids in SARS-CoV-2 viral replication and the host immune response. J Lipid Res. 2021; 62: 100129.
- Chu J, Xing C, Du Y, Duan T, Liu S et al. Pharmacological inhibition of fatty acid synthesis blocks SARS-CoV-2 replication. Nat Metab. 2021; 3: 1466-1475.
- Chukkapalli V, Heaton NS, Randall G. Lipids at the interface of virus-host interactions. Curr Opin Microbiol. 2021; 15: 512-518.
- Caterino M, Gelzo M, Sol S, Fedele R, Annunziata A et al. Dysregulation of lipid metabolism and pathological inflammation in patients with COVID-19. Sci Rep. 2021; 11: 2941.
- Zandi M, Hosseini P, Soltani S, Rasooli A, Moghadami M et al. The role of lipids in the pathophysiology of coronavirus infections. Osong Public Health Res Perspect. 2021; 12: 278-285.
- Ivanova PT, Myers DS, Milne SB, McClaren JL, Thomas PG et al. Lipid composition of viral envelope of three strains of influenza virus - not all viruses are created equal. ACS Infect Dis. 2015; 1: 399-452.
- Mingo-Casas P, Sanchez-Céspedes J, Blázquez AB, Casas J, Balsera-Manzanero M et al. Lipid signatures of West Nile virus infection unveil alterations of sphingolipid metabolism providing novel biomarkers. Emerg Microbes Infect. 2023; 12: 2231556.
- Picimbon JF. Les périrécepteurs chimiosensoriels des insectes. Med Sci. 2002; 11: 1089-1094.
- Keil TA. Sensory cilia in arthropods. Arthropod Struct Dev. 2012; 41: 515-534.
- Vogt RG. Molecular basis of pheromone detection in insects. In: Gilbert LI, Iatrou K, Gill S, eds. Comprehensive Insect Physiology, Biochemistry, Pharmacology and Molecular Biology Vol. 3, London, UK, Endocrinology, Elsevier, 2005, 753-804.
- Steiner C, Chertemps T, Maïbèche M. Diversity of biotransformation enzymes in insect antennae: possible roles in odorant activation and xenobiotic processing. In: Picimbon JF, ed. Olfactory Concepts of Insect Control-Alternative to Insecticides. Vol. 2, Springer Nature Switzerland AG, 2019, 115-145.
- Guo X, Xuan N, Liu GX, Xie HY, Lou QN, et al. An expanded survey of the moth PBP/GOBP clade in Bombyx mori: new insight into expression and functional roles. Front Physiol. 2021; 12: 712593.
- Picimbon JF. Evolution of protein physical structures in insect chemosensory systems. In: Picimbon JF, ed. Olfactory Concepts of Insect Control-Alternative to Insecticides. Vol. 2, Springer Nature Switzerland AG, 2019, 231-263.
- Sandler BH, Nikonova L, Leal WS, Clardy J. Sexual attraction in the silkworm moth: structure of the pheromone-binding protein-bombykol complex. Chem Biol. 2000; 7: 143-151.
- Lartigue A, Campanacci V, Roussel A, Larsson AM, Jones TA et al. X-ray structure and ligand binding study of a moth chemosensory protein. J Biol Chem. 2002; 277: 32094-32098.
- Xuan N, Guo X, Xie HY, Lou QN, Lu XB, et al. Increased expression of CSP and CYP genes in adult silkworm females exposed to avermectins. Insect Sci. 2015; 22: 203-219. (Insect Science Award 2017).
- Liu GX, Xuan N, Rajashekar B, Arnaud P, Offmann B et al. Comprehensive history of CSP genes: evolution, phylogenetic distribution, and functions. Genes 2020; 11: 413.
- Ishida Y. Ant antennae-specific Niemann-Pick type C2 protein. In: Picimbon JF, ed. Olfactory Concepts of Insect Control-Alternative to Insecticides. Vol. 2, Springer Nature Switzerland AG, 2019, 171-204.
- Horst R, Damberger F, Luginbühl P, Güntert P, Peng G et al. NMR structure reveals intramolecular regulation mechanism for pheromone binding and release. Proc Natl Acad Sci USA 2001; 98: 14374-14379.
- Campanacci V, Krieger J, Bette S, Sturgis JN, Lartigue A et al. Revisiting the specificity of Mamestra brassicae and Antheraea polyphemus pheromone-binding proteins with a fluorescence binding assay. J Biol Chem. 2001; 276: 20078-20084.
- Liu GX, Ma HM, Xie HY, Xuan N, Guo X et al. Biotype characterization, developmental profiling, insecticide response and binding property of Bemisia tabaci chemosensory proteins: role of CSP in insect defense. PLoS ONE 2016; 11: e0154706.
- Ishida Y, Tsuchiya W, Fujii T, Fujimoto Z, Miyazawa M et al. Niemann-Pick type C2 protein mediating chemical communication in the worker ant. Proc Natl Acad Sci USA 2014; 111: 3847-3852.
- Picimbon JF. Les protéines liant les odeurs, les protéines chimiosensorielles et les protéines Niemann-Pick type C2 des insectes contre le SARS-CoV-2 et la COVID-19: transport et dégradation des lipides de la capside virale. Insect OBPs, CSPs and NPC2s for control of SARS-CoV-2 and COVID-19: transport/degradation of viral capsid lipids. Rev Med Brux. 2023; 44: 111-122.
- Figueroa S, Freire-Paspuel B, Vega-Mariño P, Velez A, Cruz M et al. High sensitivity-low cost detection of SARS-CoV-2 by two steps end point RT-PCR with agarose gel electrophoresis visualization. Sci Rep. 2021; 11: 21658.
- Marinowic DR, Zanirati G, Rodrigues FVF, Grahl MVC, Alcará AM et al. A new SYBR Green real-time PCR to detect SARS-CoV-2. Sci Rep. 2021; 11: 2224.
- Bray N, Sopwith W, Edmunds M, Vansteenhouse H, Feenstra JDM et al. RT-PCR genotyping assays to identify SARS-CoV-2 variants in England in 2021: a design and retrospective evaluation study. Lancet Microbe 2024; 5: E173-E180.
- Liu D, Wu F, Cen Y, Ye L, Shi X et al. Comparative research on nucleocapsid and spike glycoprotein as the rapid immunodetection targets of COVID-19 and establishment of immunoassay strips. Mol Immunol. 2021; 131: 6-12.
- Cai Y, Zhang J, Xiao T, Peng H, Sterling SM et al. Distinct conformational states of SARS-CoV-2 spike protein. Science 2020; 369: 1586-1592.
- Henderson R, Edwards RJ, Mansouri K, Janowska K, Stalls V et al. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat Struct Mol Biol. 2020; 27: 925-933.
- Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARSCoV-2 entry into cells. Nat Rev Mol Cell Biol. 2022; 23: 3-20.
- Toelzer C, Gupta K, Yadav SKN, Borucu U, Davidson AD et al. Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein. Science 2020; 370: 725-730.
- Oliveira ASF, Shoemark DK, Ibarra AA, Davidson AD, Berger I et al. The fatty acid site is coupled to functional motifs in the SARSCoV-2 spike protein and modulates spike allosteric behavior. Comput Struct Biotechnol J. 2022; 20: 193-147.
- Toelzer C, Gupta K, Berger I, Schaffitzel C. Cryo-Em reveals binding of linoleic acid to SARS-CoV-2 spike glycoprotein, suggesting an antiviral treatment strategy. Acta Crystallogr D Struct Biol. 2023; 79: 111-121.
- Goc A, Sumera W, Rath M, Niedzwiecki A. Linoleic acid binds to SARS-CoV-2 RdRp and represses replication of seasonal human coronavirus OC43. Sci Rep. 2022; 12: 19114.
- Mahajan M, Chatterjee D, Bhuvaneswari K, Pillay S, Bhattacharjya S. NMR structure and localization of a large fragment of the SARS-CoV fusion protein: implications in viral cell fusion. Biochim Biophys Acta Biomembr. 2018; 1860: 407-415.
- Kawase M, Kataoka M, Shirato K, Matsuyama S. Biochemical analysis of coronavirus spike glycoprotein conformational intermediates during membrane fusion. J Virol. 2019; 93: e00785.
- Braet SM, Buckley TSC, Venkatakrishnan V, Dam KMA, Bjorkman PJ et al. Timeline of changes in spike conformational dynamics in emergent SARS-CoV-2 variants reveal progressive stabilization of trimer stalk with altered NTD dynamics. eLife 2023; 12: e82584.